bg_hom3dex¶
Background due to a homogeneous spin distribution in 3D, with excluded volume
Syntax¶
info = bg_hom3dex()
P = bg_hom3dex(t,param)
P = bg_hom3dex(t,param,lambda)
- Inputs
t– Time axis (N-array)param– Model parameterslambda– Modulation amplitude (between 0 and 1)
- Outputs
B– Model background (N-array)info– Model information (struct)
Model¶
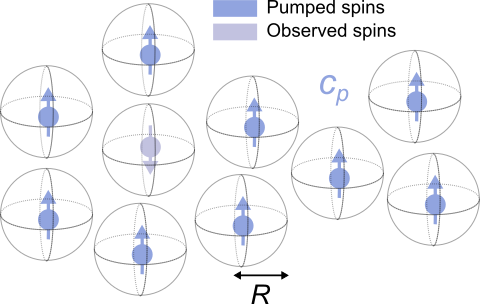
This implements a hard-shell excluded-volume model, with pumped spin concentration c (first parameter, in μM) and distance of closest approach R (second parameter, in nm).
The expression for this model is
where is the spin concentration (entered in spins/m3 into this expression) and
is the dipolar constant
The function of the exclusion distance
captures the excluded-volume effect. It is a smooth function, but doesn’t have an analytical representation. For details, see Kattnig et al, J.Phys.Chem.B 2013, 117, 16542.
| Variable | Symbol | Default | Lower bound | Upper bound | Description |
|---|---|---|---|---|---|
param(1) |
50 | 0.01 | 1000 | spin concentration (μM) | |
param(2) |
1 | 0.1 | 20 | exclusion distance (nm) |
Description¶
info = bg_hom3dex()
Returns an info structure containing the specifics of the model:
info.model– Full name of the parametric model.info.nparam– Total number of adjustable parameters.info.parameters– Structure array with information on individual parameters.
B = bg_hom3dex(t,param)
Computes the background model B from the axis t according to the parameters array param for a modulation amplitude lambda=1. The required parameters can also be found in the info structure.
B = bg_hom3dex(t,param,lambda)
Computes the background model B for a given modulation amplitude lambda (between 0 and 1).