dd_wormgauss¶
Worm-like chain model near the rigid limit with Gaussian convolution
Syntax¶
info = dd_wormgauss()
P = dd_wormgauss(r,param)
- Parameters
r- Distance axis (N-array)param- Model parameters
- Returns
P- Distance distribution (N-array)info- Model information (struct)
| Variable | Symbol | Default | Lower | Upper | Description |
|---|---|---|---|---|---|
param(1) |
3.7 | 1.5 | 10 | Contour length | |
param(2) |
10 | 2 | 100 | Persistence length | |
param(3) |
0.2 | 0.01 | 2 | Gaussian standard deviation |
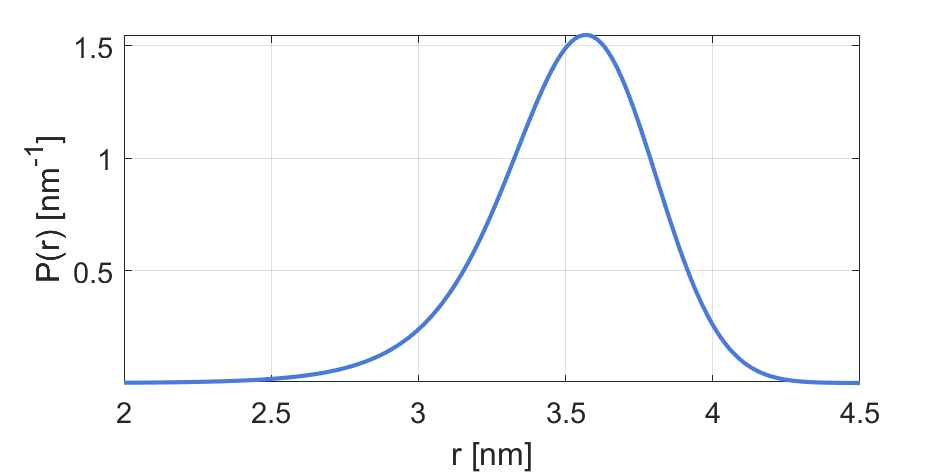
Example using default parameters:
Description¶
info = dd_wormgauss()
Returns an info structure containing the specifics of the model:
info.model- Full name of the parametric model.info.nparam- Total number of adjustable parameters.info.parameters- Structure array with information on individual parameters.
P = dd_wormgauss(r,param)
Computes the distance distribution model P from the axis r according to the parameters array param. The required parameters can also be found in the info structure.
References¶
[1] J. Wilhelm, E. Frey, Phys. Rev. Lett. 77(12), 2581-2584 (1996) DOI: 10.1103/PhysRevLett.77.2581