ex_7pdeer¶
7-pulse DEER experiment
Syntax¶
info = ex_7pdeer(t)
pathways = ex_7pdeer(t,param)
- Parameters
t- Time axis (M-array), in microsecondsparam- Model parameters (array)
- Returns
pathways- Dipolar pathways (array)info- Model information (struct)
Model¶
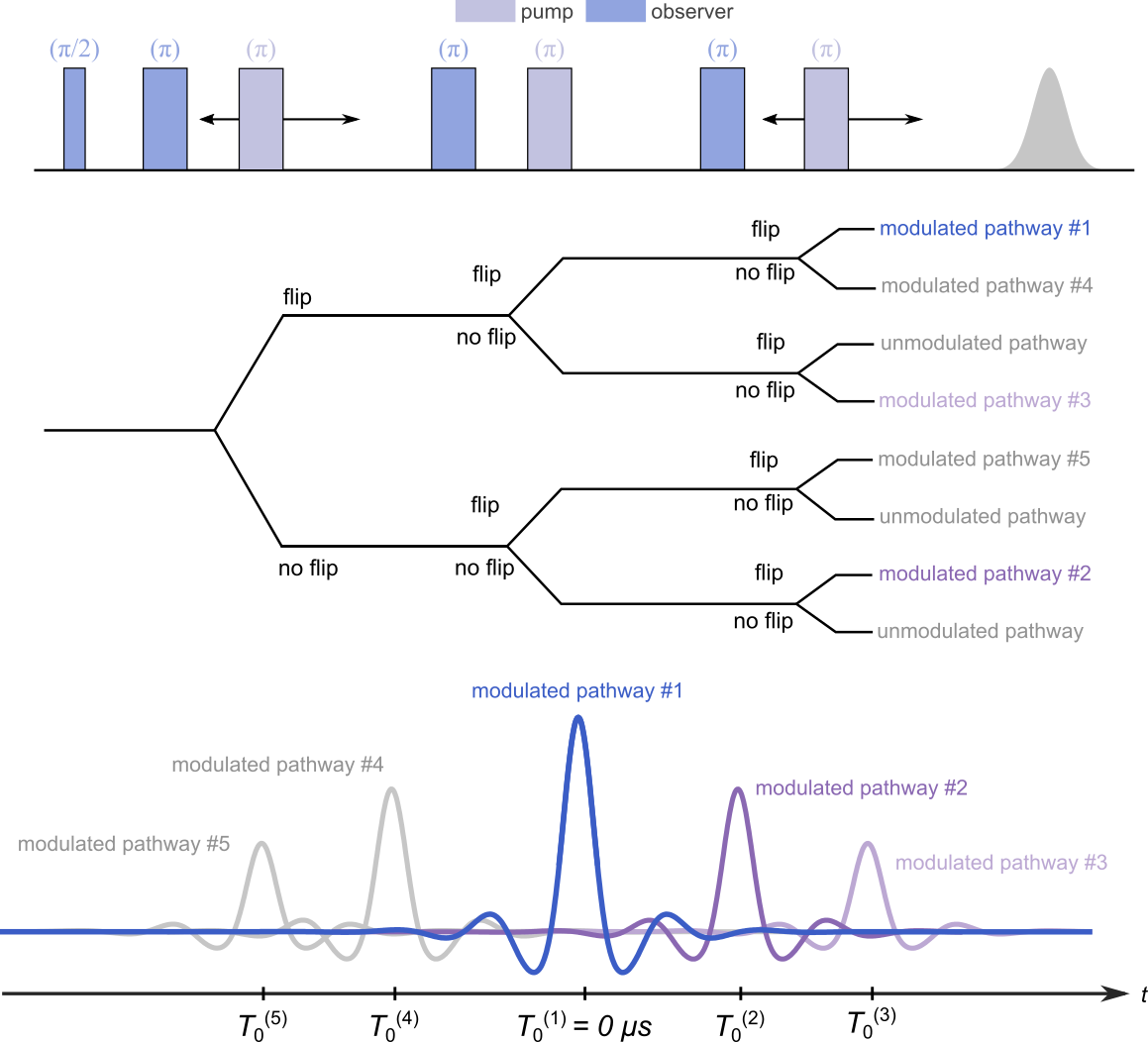
In order to reduce the parameter space, only the dipolar pathways refocusing at positive times (pathways #1-3) are considered in this model:
where ,
, and
are the refocusing times of the three modulated dipolar pathways at positive evolution times.
| Variable | Symbol | Default | Lower | Upper | Description |
|---|---|---|---|---|---|
param(1) |
0.4 | 0 | 1 | unmodulated pathways, amplitude | |
param(2) |
0.4 | 0 | 1 | 1st modulated pathway, amplitude | |
param(3) |
0.2 | 0 | 1 | 2nd modulated pathway, amplitude | |
param(4) |
0.2 | 0 | 1 | 3rd modulated pathway, amplitude | |
param(5) |
2nd modulated pathway, refocusing time (us) | ||||
param(6) |
3rd modulated pathway, refocusing time (us) |
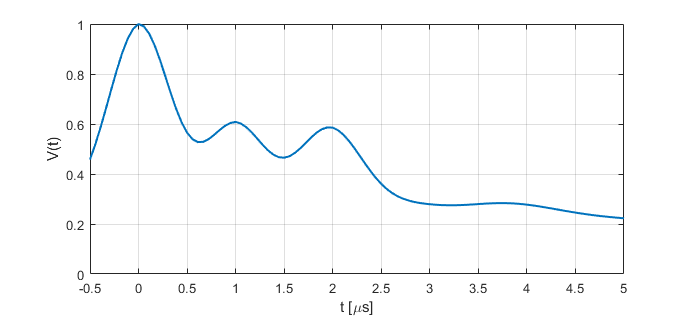
Example of a simulated signal using default parameters:
Description¶
info = ex_7pdeer(t)
Returns an info structure containing the specifics of the model:
info.model- Full name of the parametric model.info.nparam- Total number of adjustable parameters.info.parameters- Structure array with information on individual parameters.
pathways = ex_7pdeer(t,param)
Generates the dipolar pathways matrix pathways from the time-axis t and model parameters param.