deerlab.ex_4pdeer¶
- ex_4pdeer(tau1, tau2, pathways=[1, 2, 3, 4], pulselength=0.016)[source]¶
Generate a 4-pulse DEER dipolar experiment model.
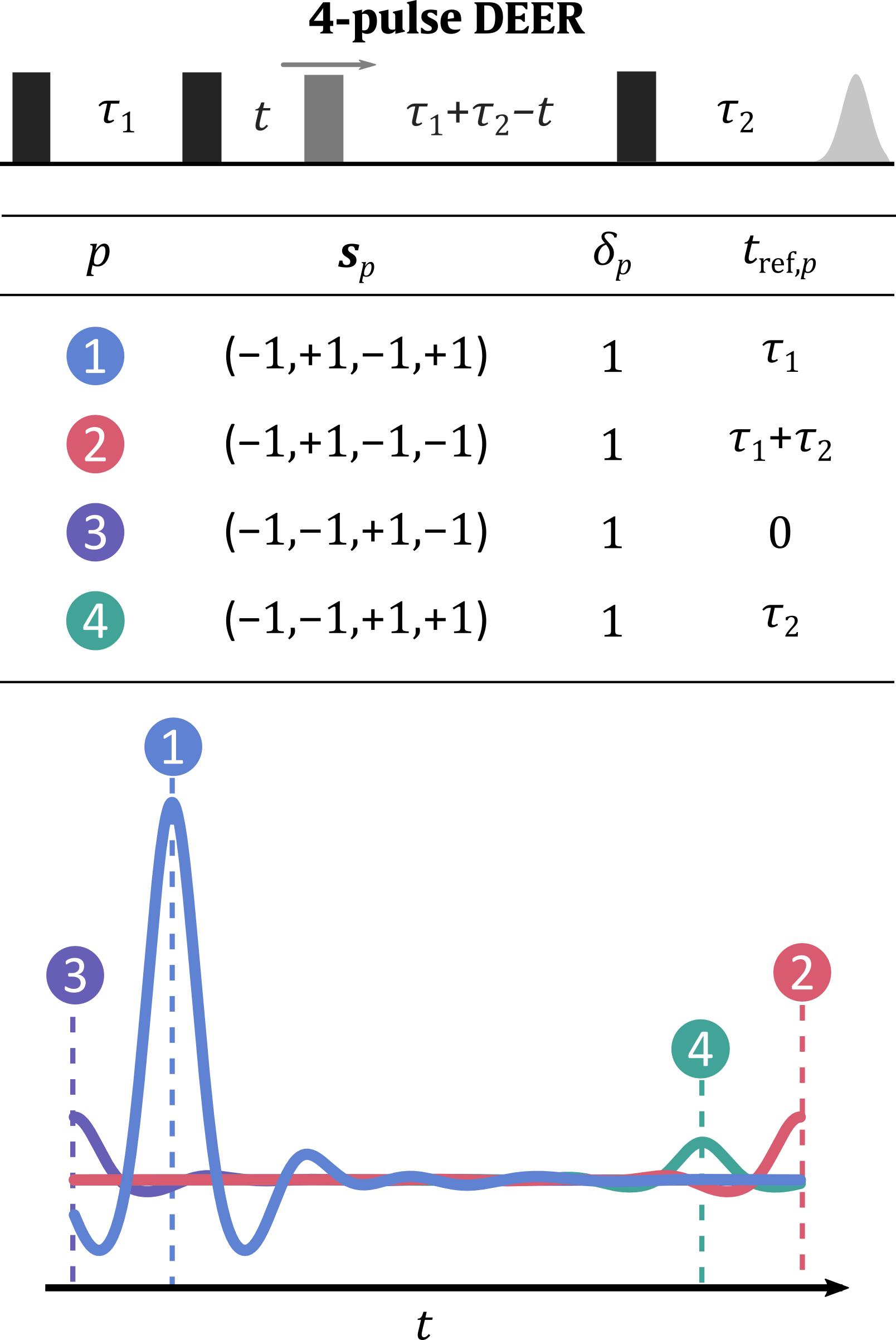
The figure below shows the dipolar pathways in 4-pulse DEER. The observer (black) and pump (grey) pulses and their interpulse delays are shown on the top. The middle table summarizes all detectable modulated dipolar pathways
along their dipolar phase accumulation factors
, harmonics
and refocusing times
. The most commonly encountered pathways are highlighted in color. The bottom panel shows a decomposition of the dipolar signal into the individual intramolecular contributions (shown as colored lines).
The dipolar time axis is defined such that
right after the second observer pulse.
Source: L. Fábregas-Ibáñez, Advanced data analysis and modeling in dipolar EPR spectroscopy, Doctoral dissertation, 2022¶
- Parameters:
- tau1float scalar
1st static interpulse delay
.
- tau2float scalar
2nd static interpulse delay
.
- pathwaysarray_like, optional
Pathways to include in the model. The pathways are specified as a list of pathways labels
. By default, pathways 1-4 are included as shown in the table above.
- pulselengthfloat scalar, optional
Length of the longest microwave pulse in the sequence in microseconds. Used to determine the uncertainty in the boundaries of the pathway refocusing times.
- Returns:
- experiment
ExperimentInfoobject Experiment object. Can be passed to
dipolarmodelto introduce better constraints into the model.
- experiment