Note
Go to the end to download the full example code.
Bootstrapped confidence intervals in routine analysis¶
How to obtain bootstrapped confidence intervals for simple routine operations.
Unless specified otherwise, the function fit will return moment-based confidence intervals based on the covariance matrix
of the objective function used to fit the data. These are quick to calculate and therefore very comfortable for quick estimates of
the uncertainty during routine analysis or testing.
However, for publication-level analysis, these confidence intervals might be inaccurate. It is strongly recommended to employ bootstrapped
confidence intervals to get accurate estimates of the uncertainty.
Conviniently, fit integrates bootstrapping to make it accessible via the keyword argument bootstrap which specifies the
number of samples to analyze to estimate the uncertainty. The larger this number, the more accurate
the confidence intervals but the longer the analysis will be. The standard for publication is typically 1000 samples.
import numpy as np
import matplotlib.pyplot as plt
import deerlab as dl
# File location
path = '../data/'
file = 'example_4pdeer_1.DTA'
# Experimental parameters
tau1 = 0.3 # First inter-pulse delay, μs
tau2 = 4.0 # Second inter-pulse delay, μs
tmin = 0.1 # Start time, μs
# Load the experimental data
t,Vexp = dl.deerload(path + file)
# Pre-processing
Vexp = dl.correctphase(Vexp) # Phase correction
Vexp = Vexp/np.max(Vexp) # Rescaling (aesthetic)
t = t - t[0] # Account for zerotime
t = t + tmin
# Distance vector
r = np.linspace(2,5,100) # nm
# Construct the model
Vmodel = dl.dipolarmodel(t,r, experiment = dl.ex_4pdeer(tau1,tau2, pathways=[1]))
# Fit the model to the data
results = dl.fit(Vmodel,Vexp,bootstrap=20)
# In this example, just for the sake of time, we will just use 20 bootstrap samples.
# Print results summary
print(results)
Bootstrap analysis with 1 cores:
0%| | 0/19 [00:00<?, ?it/s]
5%|▌ | 1/19 [00:09<02:46, 9.23s/it]
11%|█ | 2/19 [00:18<02:39, 9.39s/it]
16%|█▌ | 3/19 [00:28<02:29, 9.35s/it]
21%|██ | 4/19 [00:37<02:21, 9.42s/it]
26%|██▋ | 5/19 [00:47<02:12, 9.45s/it]
32%|███▏ | 6/19 [00:57<02:05, 9.63s/it]
37%|███▋ | 7/19 [01:06<01:53, 9.49s/it]
42%|████▏ | 8/19 [01:15<01:44, 9.50s/it]
47%|████▋ | 9/19 [01:24<01:33, 9.37s/it]
53%|█████▎ | 10/19 [01:31<01:22, 9.13s/it]
58%|█████▊ | 11/19 [01:39<01:12, 9.04s/it]
63%|██████▎ | 12/19 [01:48<01:03, 9.02s/it]
68%|██████▊ | 13/19 [01:56<00:53, 8.96s/it]
74%|███████▎ | 14/19 [02:05<00:44, 8.93s/it]
79%|███████▉ | 15/19 [02:16<00:36, 9.10s/it]
84%|████████▍ | 16/19 [02:25<00:27, 9.08s/it]
89%|████████▉ | 17/19 [02:35<00:18, 9.15s/it]
95%|█████████▍| 18/19 [02:44<00:09, 9.16s/it]
100%|██████████| 19/19 [02:53<00:00, 9.16s/it]
100%|██████████| 19/19 [02:53<00:00, 9.16s/it]
100%|██████████| 19/19 [02:53<00:00, 9.16s/it]
Goodness-of-fit:
========= ============= ============= ===================== =======
Dataset Noise level Reduced 𝛘2 Residual autocorr. RMSD
========= ============= ============= ===================== =======
#1 0.005 0.812 0.124 0.004
========= ============= ============= ===================== =======
Model hyperparameters:
==========================
Regularization parameter
==========================
0.053
==========================
Model parameters:
=========== ========= ========================= ====== ======================================
Parameter Value 95%-Confidence interval Unit Description
=========== ========= ========================= ====== ======================================
mod 0.303 (0.299,0.307) Modulation depth
reftime 0.299 (0.297,0.302) μs Refocusing time
conc 146.699 (143.846,150.507) μM Spin concentration
P ... (...,...) nm⁻¹ Non-parametric distance distribution
P_scale 1.007 (0.992,1.043) None Normalization factor of P
=========== ========= ========================= ====== ======================================
# Extract fitted dipolar signal
Vfit = results.model
Vci = results.modelUncert.ci(95)
# Extract fitted distance distribution
Pfit = results.P
Pci95 = results.PUncert.ci(95)
Pci50 = results.PUncert.ci(50)
# Extract the unmodulated contribution
Bfcn = lambda mod,conc,reftime: results.P_scale*(1-mod)*dl.bg_hom3d(t-reftime,conc,mod)
Bfit = results.evaluate(Bfcn)
Bci = results.propagate(Bfcn).ci(95)
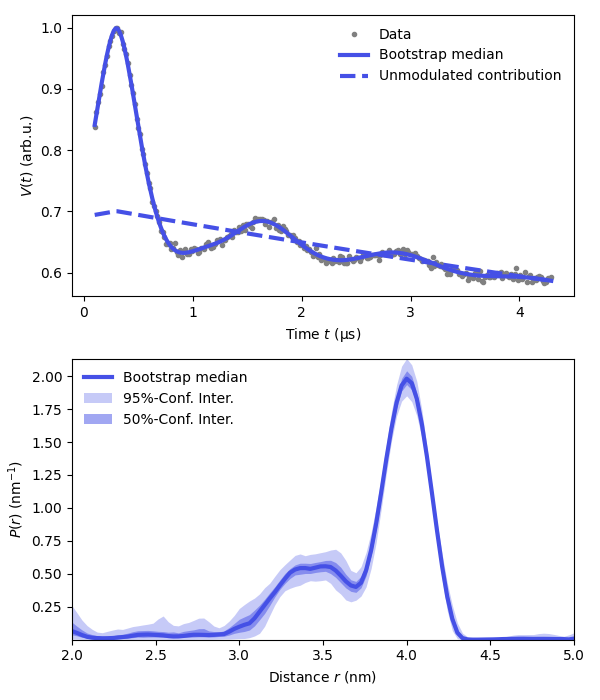
plt.figure(figsize=[6,7])
violet = '#4550e6'
plt.subplot(211)
# Plot experimental data
plt.plot(t,Vexp,'.',color='grey',label='Data')
# Plot the fitted signal
plt.plot(t,Vfit,linewidth=3,label='Bootstrap median',color=violet)
plt.fill_between(t,Vci[:,0],Vci[:,1],linewidth=0.1,label='Bootstrap median',color=violet,alpha=0.3)
plt.plot(t,Bfit,'--',linewidth=3,color=violet,label='Unmodulated contribution')
plt.legend(frameon=False,loc='best')
plt.xlabel('Time $t$ (μs)')
plt.ylabel('$V(t)$ (arb.u.)')
# Plot the distance distribution
plt.subplot(212)
plt.plot(r,Pfit,linewidth=3,label='Bootstrap median',color=violet)
plt.fill_between(r,Pci95[:,0],Pci95[:,1],alpha=0.3,color=violet,label='95%-Conf. Inter.',linewidth=0)
plt.fill_between(r,Pci50[:,0],Pci50[:,1],alpha=0.5,color=violet,label='50%-Conf. Inter.',linewidth=0)
plt.legend(frameon=False,loc='best')
plt.autoscale(enable=True, axis='both', tight=True)
plt.xlabel('Distance $r$ (nm)')
plt.ylabel('$P(r)$ (nm$^{-1}$)')
plt.tight_layout()
plt.show()
Total running time of the script: (3 minutes 14.067 seconds)