Note
Go to the end to download the full example code.
Basic analysis of a 4-pulse DEER signal¶
Fit a simple 4-pulse DEER signal with a model with a non-parametric distribution and a homogeneous background, using Tikhonov regularization.
import numpy as np
import matplotlib.pyplot as plt
import deerlab as dl
# File location
path = '../data/'
file = 'example_4pdeer_1.DTA'
# Experimental parameters
tau1 = 0.3 # First inter-pulse delay, μs
tau2 = 4.0 # Second inter-pulse delay, μs
tmin = 0.1 # Start time, μs
# Load the experimental data
t,Vexp = dl.deerload(path + file)
# Pre-processing
Vexp = dl.correctphase(Vexp) # Phase correction
Vexp = Vexp/np.max(Vexp) # Rescaling (aesthetic)
t = t - t[0] # Account for zerotime
t = t + tmin
# Distance vector
r = np.arange(2.5,5,0.01) # nm
# Construct the model
Vmodel = dl.dipolarmodel(t,r, experiment = dl.ex_4pdeer(tau1,tau2, pathways=[1]))
# Fit the model to the data
results = dl.fit(Vmodel,Vexp)
# Print results summary
print(results)
Goodness-of-fit:
========= ============= ============= ===================== =======
Dataset Noise level Reduced 𝛘2 Residual autocorr. RMSD
========= ============= ============= ===================== =======
#1 0.005 0.811 0.127 0.004
========= ============= ============= ===================== =======
Model hyperparameters:
==========================
Regularization parameter
==========================
0.224
==========================
Model parameters:
=========== ========= ========================= ====== ======================================
Parameter Value 95%-Confidence interval Unit Description
=========== ========= ========================= ====== ======================================
mod 0.302 (0.299,0.305) Modulation depth
reftime 0.299 (0.298,0.301) μs Refocusing time
conc 148.470 (143.517,153.423) μM Spin concentration
P ... (...,...) nm⁻¹ Non-parametric distance distribution
P_scale 0.999 (0.998,1.000) None Normalization factor of P
=========== ========= ========================= ====== ======================================
# Extract fitted dipolar signal
Vfit = results.model
# Extract fitted distance distribution
Pfit = results.P
Pci95 = results.PUncert.ci(95)
Pci50 = results.PUncert.ci(50)
# Extract the unmodulated contribution
Bfcn = lambda mod,conc,reftime: results.P_scale*(1-mod)*dl.bg_hom3d(t-reftime,conc,mod)
Bfit = results.evaluate(Bfcn)
Bci = results.propagate(Bfcn).ci(95)
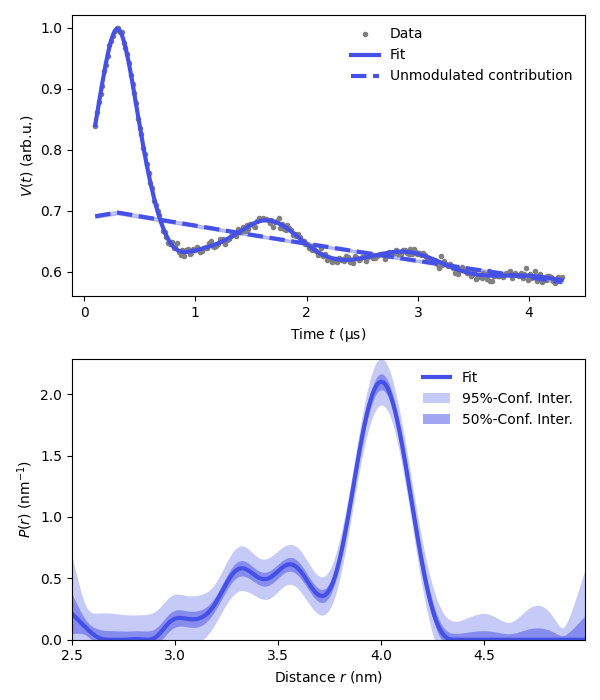
plt.figure(figsize=[6,7])
violet = '#4550e6'
plt.subplot(211)
# Plot experimental and fitted data
plt.plot(t,Vexp,'.',color='grey',label='Data')
plt.plot(t,Vfit,linewidth=3,color=violet,label='Fit')
plt.plot(t,Bfit,'--',linewidth=3,color=violet,label='Unmodulated contribution')
plt.fill_between(t,Bci[:,0],Bci[:,1],color=violet,alpha=0.3)
plt.legend(frameon=False,loc='best')
plt.xlabel('Time $t$ (μs)')
plt.ylabel('$V(t)$ (arb.u.)')
# Plot the distance distribution
plt.subplot(212)
plt.plot(r,Pfit,color=violet,linewidth=3,label='Fit')
plt.fill_between(r,Pci95[:,0],Pci95[:,1],alpha=0.3,color=violet,label='95%-Conf. Inter.',linewidth=0)
plt.fill_between(r,Pci50[:,0],Pci50[:,1],alpha=0.5,color=violet,label='50%-Conf. Inter.',linewidth=0)
plt.legend(frameon=False,loc='best')
plt.autoscale(enable=True, axis='both', tight=True)
plt.xlabel('Distance $r$ (nm)')
plt.ylabel('$P(r)$ (nm$^{-1}$)')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 17.232 seconds)