Note
Go to the end to download the full example code.
Simulating a three-pathway 4-pulse DEER signal¶
An example on how to simulate a 4-pulse DEER dipolar signal, including secondary pathways like the “2+1” contribution. This example uses a Gaussian distance distibution.
# Import the required libraries
import numpy as np
import matplotlib.pyplot as plt
import deerlab as dl
# Simulation parameters
tau1, tau2 = 0.5, 4.5 # Inter-pulse time delays, μs
tmin = 0.3 # Start time, μs
Δt = 0.008 # Time increment, μs
rmean = 4.0 # Mean distance, nm
rstd = 0.3 # Distance standard deviation, nm
Δr = 0.05 # Distance increment, nm
rmin, rmax = 1.5, 6 # Range of the distance vector, nm
conc = 50 # Spin concentration, μM
lam1 = 0.40 # Amplitude of main contribution
lam23 = 0.02 # Amplitude of the "2+1" contributions
V0 = 1 # Overall signal amplitude
# Time vector
tmax = tau1+tau2
t = np.arange(tmin, tmax, Δt)
# Distance vector
r = np.arange(rmin, rmax, Δr)
# Experiment model
experiment = dl.ex_4pdeer(tau1, tau2, pathways=[1,2,3])
# Construct the dipolar signal model
Vmodel = dl.dipolarmodel(t, r, Pmodel=dl.dd_gauss, experiment=experiment)
# Simulate the signal with orientation selection
reftime1, reftime2, reftime3 = experiment.reftimes(tau1, tau2)
Vsim = Vmodel(mean=rmean, std=rstd, conc=conc, scale=V0, lam1=lam1, lam2=lam23, lam3=lam23, reftime1=reftime1, reftime2=reftime2, reftime3=reftime3)
# Scaled background (for plotting)
Vinter = V0*(1-lam1-2*lam23)*dl.bg_hom3d(t-reftime1,conc,lam1)*dl.bg_hom3d(t-reftime2,conc,lam23)*dl.bg_hom3d(t-reftime3,conc,lam23)
# Plot the simulated signal
violet = '#4550e6'
plt.figure(figsize=[4,3])
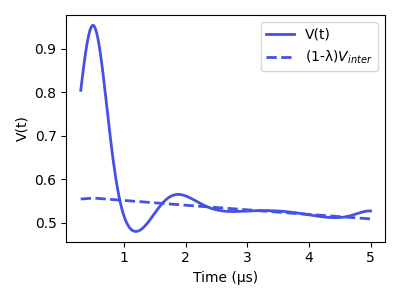
plt.plot(t, Vsim, lw=2, label='V(t)', color=violet)
plt.plot(t, Vinter, '--', color=violet, lw=2, label='(1-λ)$V_{inter}$')
plt.legend()
plt.xlabel('Time (μs)')
plt.ylabel('V(t)')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 0.109 seconds)