Note
Go to the end to download the full example code.
Dipolar pathways model selection¶
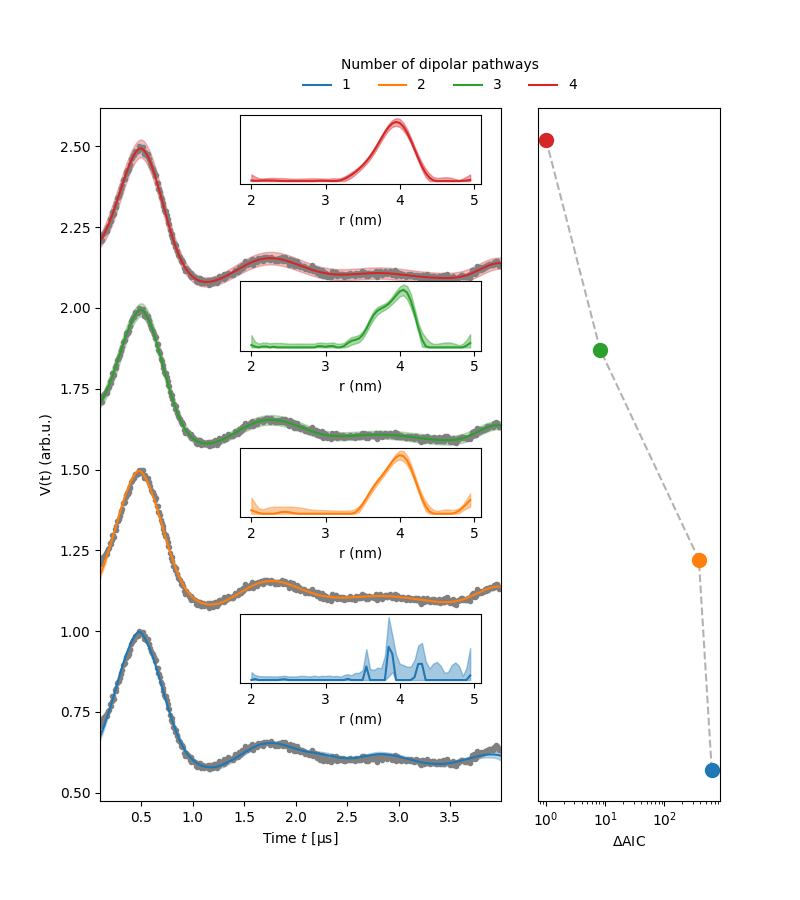
An example on how to select the number of dipolar pathways to include in the model based on the Akaike information criterion.
# Import the required libraries
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import deerlab as dl
# File location
path = '../data/'
file = 'example_4pdeer_2.DTA'
# Experimental parameters
tau1 = 0.5 # First inter-pulse delay, μs
tau2 = 3.5 # Second inter-pulse delay, μs
tmin = 0.1 # Start time, μs
# Load the experimental data
t,Vexp = dl.deerload(path + file)
# Pre-processing
Vexp = dl.correctphase(Vexp) # Phase correction
Vexp = Vexp/np.max(Vexp) # Rescaling (aesthetic)
t = t - t[0] # Account for zerotime
t = t + tmin
# Construct the distance vector
r = np.arange(2,5,0.05)
# 4-pulse DEER can have up to four different dipolar pathways
Nmax = 4
# Create the 4-pulse DEER signal models with increasing number of pathways
Vmodels = [dl.dipolarmodel(t, r, experiment=dl.ex_4pdeer(tau1,tau2,pathways=np.arange(n+1)+1)) for n in range(Nmax)]
# Fit the individual models to the data
fits = [[] for _ in range(Nmax)]
for n,Vmodel in enumerate(Vmodels):
fits[n] = dl.fit(Vmodel,Vexp)
# Extract the values of the Akaike information criterion for each fit
aic = np.array([fit.stats['aic'] for fit in fits])
# Compute the relative difference in AIC
aic = aic - aic.min() + 1 # ...add plus one for log-scale
# Plotting
colors = ['tab:blue','tab:orange','tab:green','tab:red']
fig = plt.figure(figsize=[8,9])
gs = GridSpec(1, 3, figure=fig)
ax1 = fig.add_subplot(gs[0, :-1])
for n in range(len(Vmodels)):
# Get the fits of the dipolar signal models
Vfit = fits[n].model
# Get the confidence intervals of the dipolar signal models
Vci = fits[n].propagate(Vmodels[n]).ci(95)
# Plot the experimental data
ax1.plot(t,n/2+Vexp,'.',color='grey')
# Plot the dipolar signal fits and their confidence bands
ax1.plot(t,n/2 + Vfit,label=f'{1+n}',linewidth=1.5,color=colors[n])
ax1.fill_between(t,n/2+Vci[:,0],n/2+Vci[:,1],alpha=0.3,color=colors[n])
# Plot the distance distributions as insets
for n in range(Nmax):
# Get the distance distribution and its confidence intervals
Pfit = fits[n].P
Pci = fits[n].PUncert.ci(95)
# Setup the inset plot
axins = ax1.inset_axes([0.35, 0.17+0.24*n, 0.6, 0.1])
axins.yaxis.set_ticklabels([])
axins.yaxis.set_visible(False)
# Plot the distance distributions and their confidence bands
axins.plot(r,Pfit,color=colors[n])
axins.fill_between(r,Pci[:,0],Pci[:,1],alpha=0.4,color=colors[n])
axins.set_xlabel('r (nm)')
# Plot the difference in AIC for each fit
ax2 = fig.add_subplot(gs[0,-1])
ax2.plot(aic,np.arange(Nmax),'k--',alpha=0.3)
for n in range(Nmax):
ax2.semilogx(aic[n],n,'o',markersize=10)
ax2.yaxis.set_visible(False)
# Axes settings
ax1.set_ylabel('V(t) (arb.u.)')
ax1.set_xlabel('Time $t$ [μs]')
ax1.autoscale(enable=True, axis='x', tight=True)
ax2.set_xlabel('$\Delta$AIC')
ax2.yaxis.set_ticklabels([])
# Legend settings
handles, labels = ax1.get_legend_handles_labels()
fig.legend(handles, labels, title='Number of dipolar pathways', frameon=False,
loc='upper center',ncol=Nmax,bbox_to_anchor=(0.55, 0.95))
plt.show()
Total running time of the script: (0 minutes 57.964 seconds)