Note
Go to the end to download the full example code.
Analysis of a 4-pulse DEER signal with a compactness penalty¶
Fit a simple 4-pulse DEER signal with a model with a non-parametric distribution and a homogeneous background, using Tikhonov regularization. Additionally, impose compactness of the distance distribution by penalizing for spread of the distance distribution.
import numpy as np
import matplotlib.pyplot as plt
import deerlab as dl
# File location
path = '../data/'
file = 'example_4pdeer_1.DTA'
# Experimental parameters
tau1 = 0.3 # First inter-pulse delay, μs
tau2 = 4.0 # Second inter-pulse delay, μs
tmin = 0.1 # Start time, μs
# Load the experimental data
t,Vexp = dl.deerload(path + file)
# Pre-processing
Vexp = dl.correctphase(Vexp) # Phase correction
Vexp = Vexp/np.max(Vexp) # Rescaling (aesthetic)
t = t - t[0] # Account for zerotime
t = t + tmin
# Distance vector
r = np.arange(2,5,0.025) # nm
# Construct the model
Vmodel = dl.dipolarmodel(t, r, experiment = dl.ex_4pdeer(tau1,tau2, pathways=[1]))
compactness = dl.dipolarpenalty(Pmodel=None, r=r, type='compactness')
# Fit the model to the data
results = dl.fit(Vmodel,Vexp,penalties=compactness)
# Print results summary
print(results)
Goodness-of-fit:
========= ============= ============= ===================== =======
Dataset Noise level Reduced 𝛘2 Residual autocorr. RMSD
========= ============= ============= ===================== =======
#1 0.005 0.812 0.125 0.004
========= ============= ============= ===================== =======
Model hyperparameters:
========================== ===================
Regularization parameter Penalty weight #1
========================== ===================
0.070 0.018
========================== ===================
Model parameters:
=========== ========= ========================= ====== ======================================
Parameter Value 95%-Confidence interval Unit Description
=========== ========= ========================= ====== ======================================
mod 0.302 (0.300,0.304) Modulation depth
reftime 0.299 (0.298,0.301) μs Refocusing time
conc 148.111 (144.373,151.850) μM Spin concentration
P ... (...,...) nm⁻¹ Non-parametric distance distribution
P_scale 1.000 (0.999,1.000) None Normalization factor of P
=========== ========= ========================= ====== ======================================
# Extract fitted dipolar signal
Vfit = results.model
# Extract fitted distance distribution
Pfit = results.P
Pci95 = results.PUncert.ci(95)
Pci50 = results.PUncert.ci(50)
# Extract the unmodulated contribution
Bfcn = lambda mod,conc,reftime: results.P_scale*(1-mod)*dl.bg_hom3d(t-reftime,conc,mod)
Bfit = results.evaluate(Bfcn)
Bci = results.propagate(Bfcn).ci(95)
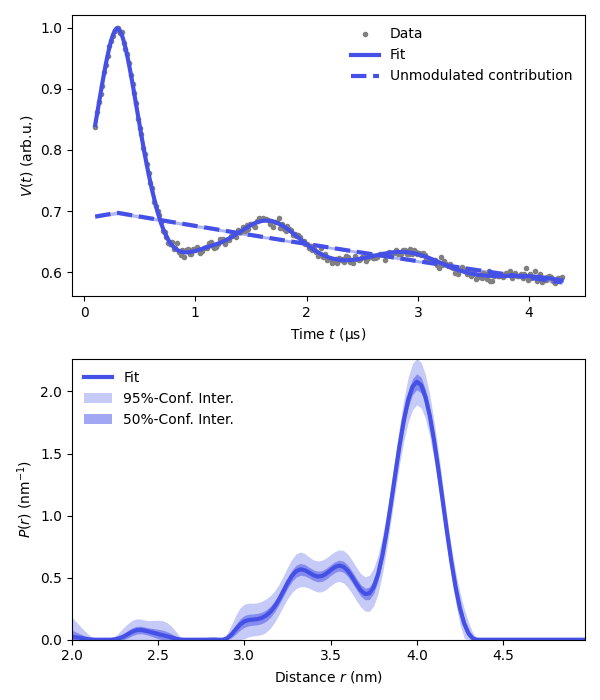
plt.figure(figsize=[6,7])
violet = '#4550e6'
plt.subplot(211)
# Plot experimental and fitted data
plt.plot(t,Vexp,'.',color='grey',label='Data')
plt.plot(t,Vfit,linewidth=3,color=violet,label='Fit')
plt.plot(t,Bfit,'--',linewidth=3,color=violet,label='Unmodulated contribution')
plt.fill_between(t,Bci[:,0],Bci[:,1],color=violet,alpha=0.3)
plt.legend(frameon=False,loc='best')
plt.xlabel('Time $t$ (μs)')
plt.ylabel('$V(t)$ (arb.u.)')
# Plot the distance distribution
plt.subplot(212)
plt.plot(r,Pfit,color=violet,linewidth=3,label='Fit')
plt.fill_between(r,Pci95[:,0],Pci95[:,1],alpha=0.3,color=violet,label='95%-Conf. Inter.',linewidth=0)
plt.fill_between(r,Pci50[:,0],Pci50[:,1],alpha=0.5,color=violet,label='50%-Conf. Inter.',linewidth=0)
plt.legend(frameon=False,loc='best')
plt.autoscale(enable=True, axis='both', tight=True)
plt.xlabel('Distance $r$ (nm)')
plt.ylabel('$P(r)$ (nm$^{-1}$)')
plt.tight_layout()
plt.show()
Total running time of the script: (2 minutes 7.723 seconds)