Note
Go to the end to download the full example code.
Fit a polynomial force field to a dipolar signal¶
This example shows how to extract a force fields (modelled as a polynomial function) from a dipolar signal.
# Import required libraries
import numpy as np
import matplotlib.pyplot as plt
import deerlab as dl
# Constants
R = 8.314 # universal gas constant, J/mol/K
T = 298 # Temperature for Boltzmann inversion (K)
kcal2J = 4.1868e3 # Conversion of kilocalories to J
thermal = R*T/kcal2J # Thermal energy in kcal/mol, (most force fields use this unit)
# Define the distance vector
r = np.linspace(2,6,300)
# Define the force field function (here modelled as a 3rd order polyomial)
def forcefield_energy(c0,c1,c2,c3):
# Evaluate polynomial model for the force-field energy
energy = np.polyval([c3,c2,c1,c0],r)
# Shift to zero energy for the minimum
energy += abs(energy.min())
return energy
def forcefield_P(c0,c1,c2,c3):
# Compute the energy
energy = forcefield_energy(c0,c1,c2,c3)
# Boltzmann distribution
Pr = np.exp(-energy/thermal)
# Ensure a probability density distribution
Pr /= np.trapezoid(Pr,r)
return Pr
# File location
path = '../data/'
file = 'example_4pdeer_4.DTA'
# Experimental parameters
tau1 = 0.3 # First inter-pulse delay, μs
tau2 = 5.0 # Second inter-pulse delay, μs
tmin = 0.1 # Start time, μs
# Load the experimental data
t,Vexp = dl.deerload(path + file)
# Pre-processing
Vexp = dl.correctphase(Vexp) # Phase correction
Vexp = Vexp/np.max(Vexp) # Rescaling (aesthetic)
t = t - t[0] # Account for zerotime
t = t + tmin
# Construct the energy and distance distribution models
forcefield_energymodel = dl.Model(forcefield_energy)
forcefield_Pmodel = dl.Model(forcefield_P)
# Set boundaries and initial conditions
forcefield_Pmodel.c1.set(lb=-4, ub=1, par0=0)
forcefield_Pmodel.c2.set(lb=-4, ub=1, par0=0)
forcefield_Pmodel.c3.set(lb=-4, ub=1, par0=0)
forcefield_Pmodel.c0.set(lb=-4, ub=1, par0=0)
# Construct the dipolar signal model
Vmodel = dl.dipolarmodel(t,r,Pmodel=forcefield_Pmodel,Bmodel=dl.bg_hom3d,experiment=dl.ex_4pdeer(tau1,tau2,pathways=[1]))
# Fit the model to the data
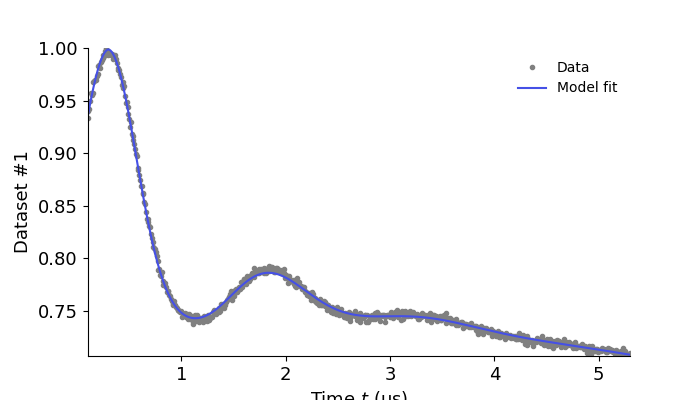
results = dl.fit(Vmodel,Vexp)
results.plot(axis=t, xlabel='Time $t$ (μs)')
plt.show()
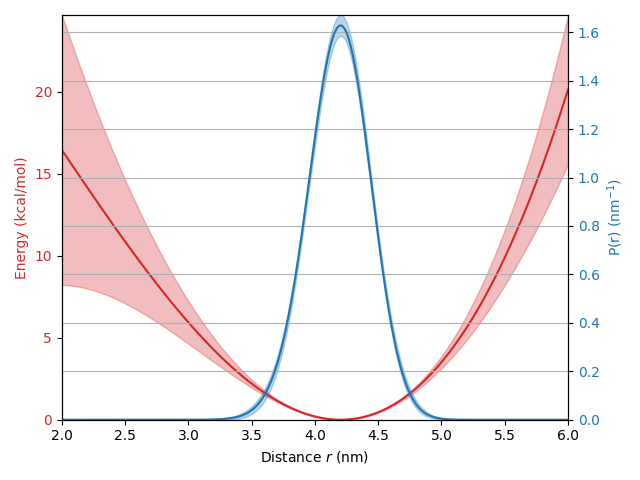
# Evaluate the models at the fitted parameters
Pfit = forcefield_Pmodel(results.c0,results.c1,results.c2,results.c3)
energy = forcefield_energymodel(results.c0,results.c1,results.c2,results.c3)
# Propagate the fit uncertainty to the models
Puq = results.propagate(forcefield_Pmodel, lb=np.zeros_like(r))
energyuq = results.propagate(forcefield_energymodel, lb=np.zeros_like(r))
# Plot the results
fig, ax1 = plt.subplots()
color = 'tab:red'
ax1.plot(r, energy, color=color)
ax1.fill_between(r,energyuq.ci(95)[:,0],energyuq.ci(95)[:,1],alpha=0.3,color=color)
ax1.tick_params(axis='y', labelcolor=color)
ax1.set_ylabel('Energy (kcal/mol)', color=color) # we already handled the x-label with ax1
ax1.set_xlabel('Distance $r$ (nm)')
ax2 = ax1.twinx() # instantiate a second axes that shares the same x-axis
color = 'tab:blue'
ax2.plot(r, Pfit, color=color)
ax2.fill_between(r,Puq.ci(95)[:,0],Puq.ci(95)[:,1],alpha=0.3,color=color)
ax2.tick_params(axis='y', labelcolor=color)
ax2.set_ylabel('P(r) (nm$^{-1}$)', color=color)
ax2.grid(None)
ax1.autoscale(enable=True, axis='both', tight=True)
ax2.autoscale(enable=True, axis='both', tight=True)
fig.tight_layout() # otherwise the right y-label is slightly clipped
plt.show()
Total running time of the script: (0 minutes 3.297 seconds)